
Open quantum systems in quantum biology
This theme has two aspects to it: theoretical physics and computational chemistry. While both address theoretical aspects of quantum biology and the use of open quantum systems, they utilise different tools.
Meet the team
Theme leaders

Professor Jim Al-Khalili
Physics lead, University of Surrey

Dr Marco Sacchi
Chemistry lead, University of Surrey
Members

Professor Dorje Brody
University of Surrey

Professor Paul Davies
Arizona State University

Dr Andrea Rocco
University of Surrey

Professor Simon Saunders
University of Oxford

Professor Cesare Tronci
University of Surrey
Research Fellows
Both areas of this theme (theoretical physics and computational chemistry) aim to address the central mystery of quantum biology: does the key to life lie in its ability to sustain long-lived quantum coherence and reversible evolution in vital biochemical processes, and how does it achieve this despite the warm and noisy cellular environment? Or is the opposite true: that life has the ability to maintain its ‘far from equilibrium’, low entropy state, which gives rise to an irreversible arrow of time?
In other words, is life ‘special’ because it can utilise the tricks of the quantum world despite the irreversibility of the arrow of time on the macroscale?
The Master Equation
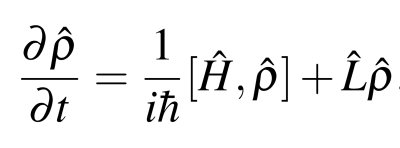
When a quantum system interacts with its surrounding environment we can no longer use the Schrödinger equation, but must solve the reduced density matrix master equation.
Research aims
We will study decoherence processes in detail to determine which features of the cellular environment prevent decoherence from happening and thereby ‘protect’ the quantum nature of the system of interest – whether that is proton tunnelling in DNA or spin coherence in green fluorescent proteins, on biologically relevant time scales.
By using an open quantum system approach we can study how a quantum system interacts with its surroundings at finite temperature, studying memory effects (i.e. non-Markovian dynamics) that arise because of the interaction of the quantum system with its environment.
We will study enzyme catalysis, capitalising on the ability of enzymes to act as highly specialised biomolecular reactors, able to perform biomolecular transformations in very variable and noisy environments, and ‘tune’ their environment to drive reactions forward by making use of quantum effects. We will quantitatively address the role of quantum tunnelling and coherence in the function of certain enzymes such as the methylamine dehydrogenase (MADH) and the effects caused by the coupling of intrinsically quantum processes through phonon and vibronic interactions.
Alongside the formal theoretical work on open quantum systems, we are also pioneering computational approaches to model biological processes using sophisticated state-of-the-art techniques such as density functional theory and multi-scale quantum mechanics/molecular dynamics.
References
- Lally, S., Werren, N., Al-Khalili, J., and Rocco, A., Master equation for non-Markovian quantum Brownian motion: The emergence of lateral coherences, Phys. Rev. A, accepted 17 Dec 2021.
- Slocombe, L., Al-Khalili, J., Sacchi, M., Quantum and classical effects in DNA point mutations: Watson- Crick tautomerism in AT and GC base pairs, Phys. Chem. Chem. Phys. 23, 4141 (2021)
- E. Mas Claret, B. Al Yahyaei, Shuyu Chu, R. M. Elliott, M. Arnaud Lopez, L. B. Meira, B.J. Howlin and D. K. Whelligan, An aza-nucleoside, fragment-like inhibitor of the DNA repair enzyme alkyladenine glycosylase (AAG), Bioorganic & Medicinal Chemistry 28, 115507 (2020)
- A Y.S. Malkhasian and B. J. Howlin, Automated drug design of kinase inhibitors to treat Chronic Myeloid Leukemia, Journal of Molecular Graphics and Modelling 19, 52-60 (2019)
- Sacchi, M.; Wales, D. J.; Jenkins, S. J., Energy landscapes and dynamics of glycine on Cu(110) Phys. Chem. Chem. Phys. 19, 16600-16605 (2017)
- D.N, Meijles, L.M, Fan, B.J. Howlin, J.M, Li, Molecular insights of p47phox phosphorylation dynamics in the regulation of NADPH oxidase activation and superoxide production, Journal of Biological Chemistry 289, 22759-22770 (2014)
- A. Dobreva, R. J. Green, I. Mueller-Harvey, Juha-Pekka Salminen ,B. J. Howlin and R. A. Frazier, Size and Molecular Flexibility Affect the Binding of Ellagitannins to Bovine Serum Albumin, M. J. Agric. Food Chem. 62(37), 9186–9194 (2014)
- Sacchi, M., Adam Y. Brewer, Stephen J. Jenkins, Julia E. Parker, Tomislav Friščić, and Stuart M. Clarke, Combined Diffraction and Density Functional Theory Calculations of Halogen-Bonded Cocrystal Monolayers, Langmuir 29, 14903-14911 (2013)
- D,N. Meijles, B.J, Howlin, J.M. Li, Consensus in silico computational modelling of the p22phox subunit of the NADPH oxidase, Computational biology and chemistry 39, 6-13 (2012)
- Sacchi, M., D. J. Wales, and S. J. Jenkins, Mode-Specific Chemisorption of CH4 on Pt{110}-(1 × 2) Explored by First-Principles Molecular Dynamics, J. Phys. Chem. C 115, 21832-21842 (2011)
- T. Blundell, D. Carney, S. Gardner ,F. Hayes, B. Howlin, T. Hubbard ,J. Overington, D. Singh, B. Sibanda and M. Sutcliffe, Knowledge‐based protein modelling and design, 18th Sir Hans Krebs Lecture, European Journal of Biochemistry 172, 513-520 (1988).