Dr Suzie Hingley-Wilson
About
Biography
I got a glimpse of life at the single cell level using my first microscope for my 10th birthday and was (Leuwen)hooked. During my Masters in Immunology at the University of Birmingham I became interested in the complex immune response to Tuberculosis (TB) and spent my PhD researching how the bacteria that causes TB, Mycobacterium tuberculosis (Mtb) can survive and live within our own immune cells which should kill them. I moved to the University of British Columbia, Canada, to continue working on this key interaction and whilst there we discovered a mechanism by which virulent Mtb can manipulate host cell death (Journal of Immunology).
To look from the side of the pathogen aswell as the host, I moved to Professor Jacob Jnr's laboratory at the Albert Einstein College of Medicine, New York to learn cutting edge mycobacterial genetics. During my time there as a Howard Hughes funded fellow we elucidated the primary attenuating deletion mechanism of BCG (published in PNAS, cited over 500 times), developed a mutant classification system (Nature Immunology) and took Bill's student's phage hunting in the Bronx zoo. Whilst in New York, I also interned at the Nature Medicine office in the Immunology and Infection section.
At Imperial College London we tried to try to uncover whether Mtb is dormant (doubtful!) and utilised patient samples to elucidate virulence mechanisms and to uncover the importance of mixed TB infections (published in Emerging Infectious Diseases and NEJM). My move to Surrey was to work in the McFadden lab on many multi-disciplinary projects before setting up my own lab working on TB. Our aim is to understand this complex human disease and to find innovative ways of curtailing this global health threat. Successful awards include the Wellcome Trust valued person award, a current multi-council AMR innovation grant and several multidisciplinary EPSRC project grants.
Affiliations and memberships
News
ResearchResearch interests
My primary research interests are tuberculosis (TB) and anti-microbial resistance (AMR). We conduct multidisciplinary research, much at the single cell level, looking at antimicrobial resistance and persistence often in the context of the host cell. We aim to identify novel mechanisms and immunotherapeutic targets by taking a global approach utilising microfluidics, mycobacterial mutagenesis, patient and environmental samples. In our lab we believe that by taking the average you often miss out the exceptional.
TB and AMR remain major global health problems. The causative agent of TB, Mycobacterium tuberculosis, can survive, replicate and persist within the toxic intracellular environment of our own immune cells and my research career has focused on elucidating the mechanisms responsible. We are currently working on how exposure to the host alters bacterial AMR and are also setting up a correlating drug screen to target and reduce AMR and TB.
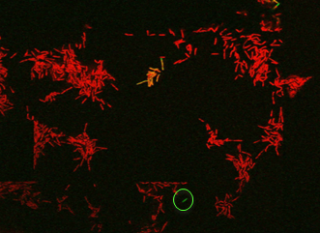
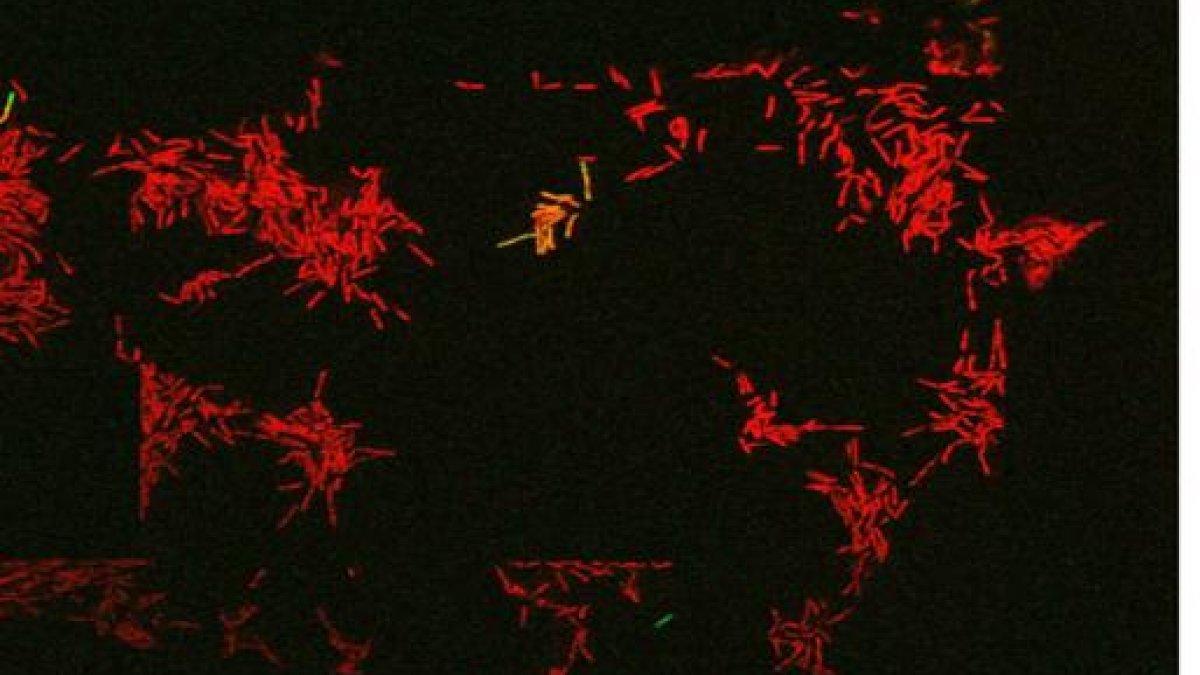
Live imaging snapshot of Mycobacterial cells treated with the front-line antibiotic rifampicin. The green GFP cell is a live persister cell which grows back upon antibiotic removal. The red cells are dead (PI stained).
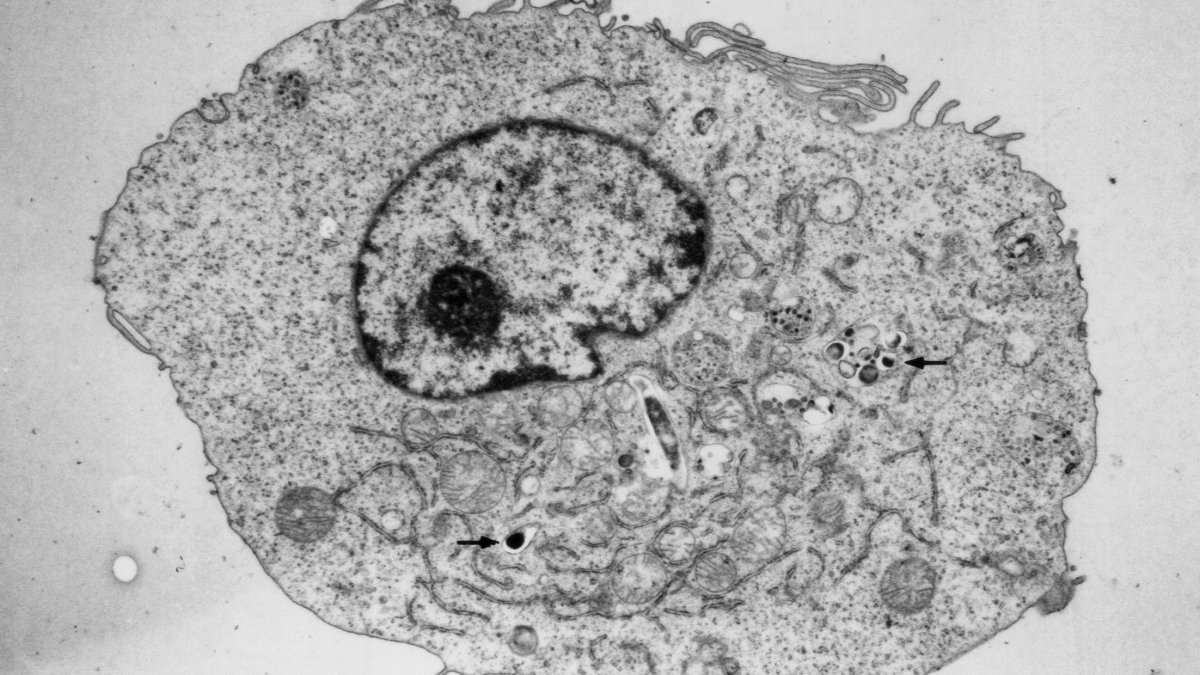
An electron micrograph of an M. tuberculosis infected macrophage. Arrows show the intracellular pathogen residing within this toxic environment.
Research interests
My primary research interests are tuberculosis (TB) and anti-microbial resistance (AMR). We conduct multidisciplinary research, much at the single cell level, looking at antimicrobial resistance and persistence often in the context of the host cell. We aim to identify novel mechanisms and immunotherapeutic targets by taking a global approach utilising microfluidics, mycobacterial mutagenesis, patient and environmental samples. In our lab we believe that by taking the average you often miss out the exceptional.
TB and AMR remain major global health problems. The causative agent of TB, Mycobacterium tuberculosis, can survive, replicate and persist within the toxic intracellular environment of our own immune cells and my research career has focused on elucidating the mechanisms responsible. We are currently working on how exposure to the host alters bacterial AMR and are also setting up a correlating drug screen to target and reduce AMR and TB.
Live imaging snapshot of Mycobacterial cells treated with the front-line antibiotic rifampicin. The green GFP cell is a live persister cell which grows back upon antibiotic removal. The red cells are dead (PI stained).

An electron micrograph of an M. tuberculosis infected macrophage. Arrows show the intracellular pathogen residing within this toxic environment.

Teaching
I teach on the bacteriology practicals (Undergraduate) and on the Masters in Medical Microbiology (including host-pathogen entry and intelligent experimental design). I also supervise undergraduate and postgraduate projects and am a Fellow of the Higher Education Academy.
Sustainable development goals
My research interests are related to the following:

Publications
Mycobacterium abscessus complex (MABC) is an important pathogen of immunocompromised patients. Accurate and rapid determination of MABC at the subspecies level is vital for optimal antibiotic therapy. Here we have used comparative genomics to design MABC subspecies-specific PCR assays. Analysis of single nucleotide polymorphisms and core genome multilocus sequence typing showed clustering of genomes into three distinct clusters representing the MABC subspecies M. abscessus, M. bolletii and M. massiliense. Pangenome analysis of 318 MABC genomes from the three subspecies allowed for the identification of 15 MABC subspecies-specific genes. In silico testing of primer sets against 1,663 publicly available MABC genomes and 66 other closely related Mycobacterium genomes showed that all assays had >97% sensitivity and >98% specificity. Subsequent experimental validation of two subspecies-specific genes each showed the PCR assays worked well in individual and multiplex format with no false-positivity with 5 other mycobacteria of clinical importance. In conclusion, we have developed a rapid, accurate, multiplex PCR-assay for discriminating MABC subspecies that could improve their detection, diagnosis and inform correct treatment choice.
Mycobacterium tuberculosis maintains long-term infections characterised by the need to regulate growth and adapt to contrasting in vivo environments. Here we show that M. tuberculosis complex bacteria utilise reversible ADP-ribosylation of single-stranded DNA as a mechanism to coordinate stationary phase growth with transcriptional adaptation. The DNA modification is controlled by DarT, an ADP-ribosyltransferase, which adds ADP-ribose to thymidine, and DarG, which enzymatically removes this base modification. Using darG-knockdown M. bovis BCG, we map the first DNA ADP-ribosylome from any organism. We show that inhibition of replication by DarT is reversible and accompanied by extensive ADP-ribosylation at the origin of replication (OriC). In addition, we observe ADP-ribosylation across the genome and demonstrate that ADP-ribose-thymidine alters the transcriptional activity of M. tuberculosis RNA polymerase. Furthermore, we demonstrate that during stationary phase, DarT-dependent ADP-ribosylation of M. tuberculosis DNA is required to optimally induce expression of the Zur regulon, including the ESX-3 secretion system and multiple alternative ribosome proteins. Thus, ADP-ribosylation of DNA can provide a mechanistic link through every aspect of DNA biology from replication to transcription to translation.
Persistence has been linked to treatment failure since its discovery over 70 years ago and understanding formation, nature and survival of this key antibiotic refractory subpopulation is crucial to enhancing treatment success and combatting the threat of antimicrobial resistance (AMR). The term ‘persistence’ is often used interchangeably with other terms such as tolerance or dormancy. In this review we focus on ‘antibiotic persistence’ which we broadly define as a feature of a subpopulation of bacterial cells that possesses the non-heritable character of surviving exposure to one or more antibiotics; and persisters as cells that possess this character. We discuss novel molecular mechanisms involved in persister cell formation, as well as environmental factors which can contribute to increased antibiotic persistence in vivo, highlighting recent developments advanced by single-cell studies. We also aim to provide a comprehensive model of persistence, the ‘hunker’ theory which is grounded in intrinsic heterogeneity of bacterial populations and a myriad of ‘hunkering down’ mechanisms which can contribute to antibiotic survival of the persister subpopulation. Finally, we discuss antibiotic persistence as a ‘stepping-stone’ to AMR and stress the urgent need to develop effective anti-persister treatment regimes to treat this highly clinically relevant bacterial sub-population.
Tuberculosis remains a leading cause of death worldwide, despite the availability of effective chemotherapy and a vaccine. Bacillus Calmette–Guérin (BCG), the tuberculosis vaccine, is an attenuated mutant of Mycobacterium bovis that was isolated after serial subcultures, yet the functional basis for this attenuation has never been elucidated. A single region (RD1), which is absent in all BCG substrains, was deleted from virulent M. bovis and Mycobacterium tuberculosis strains, and the resulting ΔRD1 mutants were significantly attenuated for virulence in both immunocompromised and immunocompetent mice. The M. tuberculosis ΔRD1 mutants were also shown to protect mice against aerosol challenge, in a similar manner to BCG. Interestingly, the ΔRD1 mutants failed to cause cytolysis of pneumocytes, a phenotype that had been previously used to distinguish virulent M. tuberculosis from BCG. A specific transposon mutation, which disrupts the Rv3874 Rv3875 (cfp-10 esat-6) operon of RD1, also caused loss of the cytolytic phenotype in both pneumocytes and macrophages. This mutation resulted in the attenuation of virulence in mice, as the result of reduced tissue invasiveness. Moreover, specific deletion of each transcriptional unit of RD1 revealed that three independent transcriptional units are required for virulence, two of which are involved in the secretion of ESAT-6 (6-kDa early secretory antigenic target). We conclude that the primary attenuating mechanism of bacillus Calmette–Guérin is the loss of cytolytic activity mediated by secreted ESAT-6, which results in reduced tissue invasiveness.
BACKGROUND: Emerging evidence suggests that Mycobacterium tuberculosis (Mtb) lineage and host ethnicity can determine tuberculosis (TB) clinical disease patterns but their relative importance and interaction are unknown. METHODS: We evaluated prospectively collected TB surveillance and Mtb strain typing data in an ethnically heterogeneous UK population. Lineage assignment was denoted using 15-loci mycobacterial interspersed repetitive units containing variable numbers of tandem repeats (MIRU-VNTR) and MIRU-VNTRplus. Geographical and ethnic associations of the six global Mtb lineages were identified and the influence of lineage and demographic factors on clinical phenotype were assessed using multivariate logistic regression. RESULTS: Data were available for 1070 individuals with active TB which was pulmonary only, extrapulmonary only and concurrent pulmonary-extrapulmonary in 52.1%, 36.9% and 11.0% respectively. The most prevalent lineages were Euro-American (43.7%), East African Indian (30.2%), Indo-Oceanic (13.6%) and East Asian (12.2%) and were geo-ethnically restricted with, for example, Indian subcontinent ethnicity inversely associated with Euro-American lineage (OR 0.23; 95% CI 0.14 to 0.39) and positively associated with the East African-Indian lineage (OR 4.04; 95% CI 2.19 to 7.45). Disease phenotype was most strongly associated with ethnicity (OR for extrathoracic disease 21.14 (95% CI 6.08 to 73.48) for Indian subcontinent and 14.05 (3.97 to 49.65) for Afro-Caribbean), after adjusting for lineage. With East Asian lineage as the reference category, the Euro-American (OR 0.54; 95% CI 0.32 to 0.91) and East-African Indian (OR 0.50; 95% CI 0.29 to 0.86) lineages were negatively associated with extrathoracic disease, compared with pulmonary disease, after adjusting for ethnicity. CONCLUSIONS: Ethnicity is a powerful determinant of clinical TB phenotype independently of mycobacterial lineage and the role of ethnicity-associated factors in pathogenesis warrants investigation.
The analysis of cell electrophysiology for pathogenic samples at BSL3 can be problematic. It is virtually impossible to isolate infected from uninfected without a label, for example green fluorescent protein, which can potentially alter the cell electrical properties. Furthermore, the measurement of highly pathogenic organisms often requires equipment dedicated only for use with these organisms due to safety considerations. To address this, we have used dielectrophoresis to study the electrical properties of the human THP-1 cell line and monocyte-derived macrophages before and after infection with non-labelled Mycobacterium tuberculosis. Infection with these highly pathogenic bacilli resulted in changes including a raised surface conductance (associated with reduced zeta potential) and increased capacitance, suggesting an increase in surface roughness. We have also investigated the effect of fixation on THP-1 cells as a means to enable study on fixed samples in BSL1 or 2 laboratories, which suggests that the properties of these cells are largely unaffected by the fixation process. This advance results in a novel technique enabling the isolation of infected and non-infected cells in a sample without labelling.
Persistence, the phenomenon whereby a small subpopulation of bacterial cells survive sterilization, prolongs antibiotic treatment and contributes to the development of genetic antimicrobial drug resistance (AMR). In this study we performed single-cell tracking of wild-type and high-persister mutant strains of Escherichia coli to identify factors that correlate with persistence. We found, as expected, persistence correlated with slow growth, but also with small birth size. We investigated intergenerational (mother–daughter) and intragenerational (sister–sister) phenotypic inheritance of growth parameters and discovered the mutant phenotype was associated with lower levels of phenotypic inheritance and identified the gene responsible, the transcription factor ydcI. Targeting pathways involved in persistence could reveal approaches to impeding persistence and the development of AMR.
The clinical, radiological and pathological similarities between sarcoidosis and tuberculosis can make disease differentiation challenging. A complicating factor is that some cases of sarcoidosis may be initiated by mycobacteria. We hypothesised that immunological profiling might provide insight into a possible relationship between the diseases or allow us to distinguish between them.
Whenever a genetically homogenous population of bacterial cells is exposed to antibiotics, a tiny fraction of cells survives the treatment,the phenomenon known as bacterial persistence [G.L. Hobby et al., Exp. Biol. Med. 50, 281–285 (1942); J. Bigger, The Lancet 244, 497– 500 (1944)]. Despite its biomedical relevance, the origin of the phenomenon is still unknown, and as a rare, phenotypically resistant subpopulation, persisters are notoriously hard to study and define. Using computerized tracking we show that persisters are small at birth and slowly replicating. We also determine that the highpersister mutant strain of Escherichia coli, HipQ, is associated with the phenotype of reduced phenotypic inheritance (RPI). We identify the gene responsible for RPI, ydcI, which encodes a transcription factor, and propose a mechanism whereby loss of phenotypic inheritance causes increased frequency of persisters. These results provide insight into the generation and maintenance of phenotypic variation and provide potential targets for the development of therapeutic strategies that tackle persistence in bacterial infections.
The 6-kDa early secretory antigenic target of Mycobacterium tuberculosis (ESAT-6) and the 10-kDa culture filtrate antigen (CFP-10), encoded in region of difference 1 (RD1) and secreted by the ESAT-6 system 1 (Esx-1) secretion system, are the most immunodominant and highly M. tuberculosis (MTB)-specific antigens. These attributes are responsible for their primary importance in tuberculosis (TB) immunodiagnosis and vaccine development. Rv3615c [Esx-1 substrate protein C (EspC)], encoded outside RD1, is similar in size and sequence homology to CFP-10 and ESAT-6, suggesting it might be a target of cellular immunity in TB. Using ex vivo enzyme-linked immunospot- and flow cytometry-based cytokine-secretion assay, we comprehensively assessed cellular immune responses to EspC in patients with active TB, latently infected persons, and uninfected bacillus Calmette-Guérin (BCG)-vaccinated controls. EspC was at least as immunodominant as ESAT-6 and CFP-10 in both active and latent TB infection. EspC contained broadly recognized CD4(+) and CD8(+) epitopes, inducing a predominantly CD4(+) T-cell response that comprised functional T-cell subsets secreting both IFN-γ and IL-2 as well as functional T-cell subsets secreting only IFN-γ. Surprisingly, T-cell responses to EspC were as highly specific (93%) for MTB infection as responses to ESAT-6 and CFP-10, with only 2 of 27 BCG-vaccinated controls responding to each antigen. Using quantitative proteomics and metabolically labeled mutant and genetically complemented MTB strains, we identified the mechanism of the specificity of anti-EspC immunity as the Esx-1 dependence of EspC secretion. The high immunodominance of EspC, equivalent to that of ESAT-6 and CFP-10, makes it a TB vaccine candidate, and its high specificity confers strong potential for T-cell-based immunodiagnosis.
In low-resource settings with high tuberculosis (TB) burdens, lack of rapid diagnostic methods for detection and differentiation of complex (MTBC) is a major challenge affecting TB management. This study utilized comparative genomic analyses of MTBC lineages; , Lineages 5/6 and to identify lineage-specific genes. Primers were designed for the development of a Multiplex PCR assay which was successful in differentiating the MTBC lineages. There was no cross-reaction with other respiratory pathogens tested. Validation of the assay using clinical samples was performed with sputum DNA extracts from 341 clinically confirmed active TB patients. It was observed that 24.9% of cases were caused by , while L5 & L6 reported 9.0% and 14.4%, respectively. infection was the least frequently detected lineage with 1.8%. Also, 27.0% and 17.0% of the cases were PCR negative and unspeciated, respectively. However, mixed-lineage TB infections were recorded at a surprising 5.9%. This multiplex PCR assay will allow speciation of MTBC lineages in low-resource regions, providing rapid differentiation of TB infections to select appropriate medication at the earliest possible time point. It will also be useful in epidemiological surveillance studies providing reliable information on the prevalence of TB lineages as well as identifying difficult to treat cases of mixed-lineage tuberculosis infections.
It is thought that during latent infection, Mycobacterium tuberculosis bacilli are retained within granulomas in a low-oxygen environment. The dormancy survival (Dos) regulon, regulated by the response regulator DosR, appears to be essential for hypoxic survival in M. tuberculosis, but it is not known how the regulon promotes survival. Here we report that mycobacteria, in contrast to enteric bacteria, do not form higher-order structures (e.g. ribosomal dimers) upon entry into stasis. Instead, ribosomes are stabilized in the associated form (70S). Using a strategy incorporating microfluidic, proteomic, and ribosomal profiling techniques to elucidate the fate of mycobacterial ribosomes during hypoxic stasis, we show that the dormancy regulator DosR is required for optimal ribosome stabilization. We present evidence that the majority of this effect is mediated by the DosR-regulated protein MSMEG_3935 (a S30AE domain protein), which is associated with the ribosome under hypoxic conditions. A Δ3935 mutant phenocopies the ΔdosR mutant during hypoxia, and complementation of ΔdosR with the MSMEG_3935 gene leads to complete recovery of dosR mutant phenotypes during hypoxia. We suggest that this protein is named ribosome-associated factor under hypoxia (RafH) and that it is the major factor responsible for DosR-mediated hypoxic survival in mycobacteria.
As the goal of a bacterium is to become bacteria, evolution has imposed continued selections for gene expression. The intracellular pathogen , the causative agent of tuberculosis, has adopted a fine-tuned response to survive its host's methods to aggressively eradicate invaders. The development of microarrays and later RNA sequencing has led to a better understanding of biological processes controlling the relationship between host and pathogens. In this study, RNA-seq was performed to detail the transcriptomes of grown in various conditions related to stresses endured by during host infection and to delineate a general stress response incurring during persisting macrophage stresses. was subjected to long-term growth, nutrient starvation, hypoxic and acidic environments. The commonalities between these stresses point to maneuvering to exploit propionate metabolism for lipid synthesis or to withstand propionate toxicity whilst in the intracellular environment. While nearly all stresses led to a general shutdown of most biological processes, up-regulation of pathways involved in the synthesis of amino acids, cofactors, and lipids were observed only in hypoxic . This data reveals genes and gene cohorts that are specifically or exclusively induced during all of these persisting stresses. Such knowledge could be used to design novel drug targets or to define possible vulnerabilities for vaccine development. Furthermore, the disruption of specific functions from this gene set will enhance our understanding of the evolutionary forces that have caused the tubercle bacillus to be a highly successful pathogen.
Tuberculosis (TB) is a global health problem caused by the airborne pathogen Mycobacterium tuberculosis. Currently, one‐third of the world's population is infected with M. tuberculosis, and this slow, tenacious bacterium kills 1.6 million people around the world each year, equating to over 4,300 deaths every day (1). Failure to eradicate this age‐old disease is the result of an ineffective vaccine and extended, often insufficient, chemotherapy. To date, the only licensed vaccine available is Mycobacterium bovis BCG, a live attenuated strain of M. bovis discovered in 1919 by Albert Calmette and Camille Guérin following 230 subcultures of the original virulent isolate (2, 3). Distribution of this vaccine to various countries, and more subculturing, led to genetic variations between different BCG strains. However, all strains possess a common deletion that occurred prior to 1919. The deleted region is called region of difference 1 (RD1), and it encodes a key part of the type VII secretion system known as ESAT‐6 secretion system 1 (ESX‐1) (Fig. 1A); deletion(s) in this particular region are considered the major cause of BCG attenuation (4–6).
A biocoating confines non-growing, metabolically-active bacteria within a synthetic colloidal polymer (i.e. latex) film. Bacteria encapsulated inside biocoatings can perform useful functions, such as a biocatalyst in wastewater treatment. A biocoating needs to have high a permeability to allow a high rate of mass transfer for rehydration and the transport of both nutrients and metabolic products. It therefore requires an interconnected porous structure. Tuning the porosity architecture is a challenge. Here, we exploited rigid tubular nanoclays (halloysite) and non-toxic latex particles (with a relatively high glass transition temperature) as the colloidal “building blocks” to tailor the porosity inside biocoatings containing Escherichia coli bacteria as a model organism. Electron microscope images revealed inefficient packing of the rigid nanotubes and proved the existence of nanovoids along the halloysite/polymer interfaces. Single-cell observations using confocal laser scanning microscopy provided evidence for metabolic activity of the E. coli within the biocoatings through the expression of yellow fluorescent protein. A custom-built apparatus was used to measure the permeability of a fluorescein sodium salt in the biocoatings. Whereas there was no measurable permeability in a coating made from only latex particles, the permeability coefficient of the composite biocoatings increased with increasing halloysite content up to a value of 110-4 m h-1. The effects of this increase in permeability was demonstrated through a specially-developed resazurin reduction assay. Bacteria encapsulated in halloysite composite biocoatings had statistically significant higher metabolic activities in comparison to bacteria encapsulated in a non-optimized coating made from latex particles alone.
Antibiotic persistence is a phenomenon observed when genetically susceptible cells survive long-term exposure to antibiotics. These 'persisters' are an intrinsic component of bacterial populations and stem from phenotypic heterogeneity. Persistence to antibiotics is a concern for public health globally, as it increases treatment duration and can contribute to treatment failure. Furthermore, there is a growing array of evidence that persistence is a 'stepping-stone' for the development of genetic antimicrobial resistance. Urinary tract infections (UTIs) are a major contributor to antibiotic consumption worldwide, and are known to be both persistent (i.e. affecting the host for a prolonged period) and recurring. Currently, in clinical settings, routine laboratory screening of pathogenic isolates does not determine the presence or the frequency of persister cells. Furthermore, the majority of research undertaken on antibiotic persistence has been done on lab-adapted bacterial strains. In the study presented here, we characterized antibiotic persisters in a panel of clinical uropathogenic Escherichia coli isolates collected from hospitals in the UK and Australia. We found that a urine-pH mimicking environment not only induces higher levels of antibiotic persistence to meropenem and colistin than standard laboratory growth conditions, but also results in rapid development of transient colistin resistance, regardless of the genetic resistance profile of the isolate. Furthermore, we provide evidence for the presence of multiple virulence factors involved in stress resistance and biofilm formation in the genomes of these isolates, whose activities have been previously shown to contribute to the formation of persister cells.
Mcl-1 protein expression was found to be up-regulated during infection with virulent Mycobacterium tuberculosis strain H37Rv. Mcl-1 induction in THP-1 cells was optimal at a multiplicity of infection of 0.8–1.2 bacilli per macrophage and was independent of opsonin coating of the bacteria. Mcl-1 expression was elevated as early as 4 h, peaked at 5.8-fold above control cells at 24 h, and remained elevated at 48 h after infection. In THP-1 cells, mMcl-1 mRNA was induced by infection with live H37Rv but not with attenuated M. tuberculosis strain H37Ra, heat-killed H37Rv, or latex beads. In THP-1 cells and monocyte-derived macrophages (MDMs), Mcl-1 protein was induced by infection with live H37Rv but not with attenuated M. tuberculosis strain H37Ra, heat-killed H37Rv, or latex beads. Treatment of uninfected, H37Ra-infected, and H37Rv-infected THP-1 cells and MDMs with antisense oligonucleotides to mcl-1 reduced Mcl-1 expression by >84%. This resulted in an increase in apoptosis of both MDMs and THP-1 cells that were infected with H37Rv, but not cells that were uninfected or infected with H37Ra. Increased apoptosis correlated with a decrease in M. tuberculosis CFUs recovered from antisense-treated, H37Rv-infected cells at 4 and 7 days after infection. In contrast, CFU recoveries from sense-treated, H37Rv-infected cells or from antisense- or sense-treated, H37Ra-infected cells were unchanged from controls. Thus, the antiapoptotic effect of the induction of Mcl-1 expression in H37Rv-infected macrophages promotes the survival of virulent M. tuberculosis.
Hydrogen offers a source of energy that does not produce any greenhouse gas when combusted. However, some manufacturing methods of hydrogen consume large amounts of energy and produce carbon dioxide as a by-product. The production of hydrogen by bacteria is an attractive alternative, because it is not energy intensive and - under the right conditions - does not release greenhouse gases. In this review, we introduce the five known ways by which bacteria can evolve hydrogen. We then describe methods to encapsulate living bacteria in synthetic layers, called coatings, for applications in bioreactors. We review the few examples in which biocoatings have been used to produce hydrogen via the photo-fermentation method. Although not used in biocoatings so far, the dark fermentation method of hydrogen production avoids the need for illumination while offering a high yield with low oxygen evolution. We identify the potential for using genetically-modified bacteria in future research on biocoatings.
ABSTRACT Biocoatings, in which viable bacteria are immobilized within a waterborne polymer coating for a wide range of potential applications, have garnered greater interest in recent years. In bioreactors, biocoatings can be ready-to-use alternatives for carbon capture or biofuel production that could be reused multiple times. Here, we have immobilized cyanobacteria in mechanically hard biocoatings, which were deposited from polymer colloids in water (i.e ., latex). The biocoatings are formed upon heating to 37°C and fully dried before rehydrating. The viability and oxygen evolution of three cyanobacterial species within the biocoatings were compared. Synechococcus sp. PCC 7002 was non-viable inside the biocoatings immediately after drying, whereas Synechocystis sp. PCC 6803 survived the coating formation, as shown by an adenosine triphosphate (ATP) assay. Synechocystis sp. PCC 6803 consumed oxygen (by cell respiration) for up to 5 days, but was unable to perform photosynthesis, as indicated by a lack of oxygen evolution. However, Chroococcidiopsis cubana PCC 7433, a strain of desiccation-resistant extremophilic cyanobacteria, survived and performed photosynthesis and carbon capture within the biocoating, with specific rates of oxygen evolution up to 0.4 g of oxygen/g of biomass per day. Continuous measurements of dissolved oxygen were carried out over a month and showed no sign of decreasing activity. Extremophilic cyanobacteria are viable in a variety of environments, making them ideal candidates for use in biocoatings and other biotechnology. IMPORTANCE As water has become a precious resource, there is a growing need for less water-intensive use of microorganisms, while avoiding desiccation stress. Mechanically robust, ready-to-use biocoatings or “living paints” (a type of artificial biofilm consisting of a synthetic matrix containing functional bacteria) represent a novel way to address these issues. Here, we describe the revolutionary, first-ever use of an extremophilic cyanobacterium ( Chroococcidiopsis cubana PCC 7433) in biocoatings, which were able to produce high levels of oxygen and carbon capture for at least 1 month despite complete desiccation and subsequent rehydration. Beyond culturing viable bacteria with reduced water resources, this pioneering use of extremophiles in biocoatings could be further developed for a variety of applications, including carbon capture, wastewater treatment and biofuel production. As water has become a precious resource, there is a growing need for less water-intensive use of microorganisms, while avoiding desiccation stress. Mechanically robust, ready-to-use biocoatings or “living paints” (a type of artificial biofilm consisting of a synthetic matrix containing functional bacteria) represent a novel way to address these issues. Here, we describe the revolutionary, first-ever use of an extremophilic cyanobacterium ( Chroococcidiopsis cubana PCC 7433) in biocoatings, which were able to produce high levels of oxygen and carbon capture for at least 1 month despite complete desiccation and subsequent rehydration. Beyond culturing viable bacteria with reduced water resources, this pioneering use of extremophiles in biocoatings could be further developed for a variety of applications, including carbon capture, wastewater treatment and biofuel production.
Infections with >1 Mycobacterium tuberculosis strain(s) are underrecognized. We show, in vitro and in vivo, how first-line treatment conferred a competitive growth advantage to amplify a multidrug-resistant M. tuberculosis strain in a patient with mixed infection. Diagnostic techniques that identify mixed tubercle bacilli populations are needed to curb the spread of multidrug resistance.
Studying defined mutants of Mycobacterium tuberculosis in the mouse model of infection has led to the discovery of attenuated mutants that fall into several phenotypic classes. These mutants are categorized by their growth characteristics compared with those of wild-type M. tuberculosis, and include severe growth in vivo mutants, growth in vivo mutants, persistence mutants, pathology mutants and dissemination mutants. Here, examples of each of these mutant phenotypes are described and classified accordingly. Defining the importance of mycobacterial gene products responsible for in vivo growth, persistence and the induction of immunopathology will lead to a greater understanding of the host-pathogen interaction and potentially to new antimycobacterial treatment options.
BackgroundThe use of motor tricycles in transporting municipal solid waste (MSW) within urban and peri-urban towns in Ghana is on the increase. This activity often leads to the introduction of pathogen-containing bioaerosols into the environment, as well as to the tricycle operators. We sought to investigate the prevalence and associated risk factors of respiratory pathogens among solid waste tricycle operators. MethodsA cross-sectional study was conducted among 155 solid waste transporters who use motor tricycles using semi-structured interviews. Nasopharyngeal swabs were obtained from participants and screened for respiratory pathogens using Polymerase Chain Reaction (PCR). ResultsPathogens detected in participants were SARS-CoV-2 (n = 10, 6.5%) and Streptococcus pneumoniae (n = 10, 6.5%), constituting an overall prevalence of 12.9% and co-infection rate of 1.3%. The most common self-reported symptoms were cough (n = 67, 43.2%), sore throat (n = 44, 28.4%) and difficulty in breathing (n = 22, 14.2%). Adherence to the use of gloves (n = 117, 75.5%) and nose mask (n = 110, 71.0%) was high. There was a significant association between the detection of respiratory pathogens and the use of gloves, use of more than one PPE and exposure to other pollutants (p < 0.05). Individuals who were exposed to "other pollutants" significantly had lower odds of becoming infected with respiratory pathogens (Adj. OR (95% CI): 0.119(0.015,0.938). ConclusionAlthough prevalence of respiratory pathogens is generally low, strict adherence to PPE use could further reduce its rates to even lower levels. Governmental health institutions and informal solid waste transporters should address challenges related to exposure to pollutants, use of gloves, and multiple PPE.
Non-human primate models of Tuberculosis (TB) are one of the most commonly used within the experimental TB field because they closely mimic the whole spectrum of disease progression of human TB. However, the early cellular interactions of the pulmonary granuloma are still not well understood. The use of this model allows investigation into the early interactions of cells within pulmonary granulomas which cannot be undertaken in human samples. Pulmonary granulomas from rhesus and cynomolgus macaques from two timepoints post infection were categorised into categories 1 - 6 (early to late stage granulomas) and immunohistochemistry was used to identify CD68+ macrophages, CD3+ T cells and CD20+ B cells. Multinucleated giant cells and acid-fast bacilli were also quantified. At week four post infection, cynomolgus macaques were found to have more CD68+ cells than rhesus in all but category 1 granulomas. Cynomolgus also had a significantly higher percentage of CD20+ B cells in category 1 granulomas. At week twelve post infection, CD68+ cells were most abundant in category 4 and 5 granulomas in both species; however, there were no significant differences between them. CD3+ T cells and CD20+ B cells were significantly higher in the majority of granuloma categories in cynomolgus compared to rhesus. Multinucleated giant cells and acid-fast bacilli were most abundant in categories 5 and 6 at week 12 post challenge in both species. This study has identified the basic cellular composition and spatial distribution of immune cells within pulmonary granulomas in both rhesus and cynomolgus macaques over time. The data from this study will add to the knowledge already gained in this field and may inform future research on vaccines and therapeutics for TB.
With the completion of genome sequencing of Mycobacterium tuberculosis and upsurge in the incidence of M. tuberculosis infection worldwide partly as a result of HIV pandemic, there is need for rationale approach to vaccine and chemotherapy discoveries for M. tuberculosis. The homologue of mig gene of Mycobacterium avium was searched for in the M. tuberculosis database at The Institute of Genomic Research (TIGR), USA and The Sanger Institute, UK. Homologue of the gene was found and comprehensively analysed. Reverse transcription PCR (RT-PCR) was carried out on the mig (fadD19) gene homologue and echA19 gene. The result of the RT-PCR showed that the mig gene was at least 2-fold upregulated during intracellular infection of macrophage compared to the broth grown bacilli as opposed to the demonstrated specific expression of mig gene in M. avium infected macrophage. The echA19 gene was also found to be upregulated. . © Ibadan Biomedical Communications Group.
Cell growth experiments with a microfluidic device produce large scale time-lapse image data, which contain important information on cell growth and patterns in their genealogy. To extract such information, we propose a scheme to segment and track bacterial cells automatically. In contrast to most published approaches, which often split segmentation and tracking into two independent procedures, we focus on designing an algorithm that describes cell properties evolving between consecutive frames by feeding segmentation and tracking results from one frame to the next one. The cell boundaries are extracted by minimising the Distance Regularised Level Set Evolution model. Each individual cell was identified and tracked by identifying cell septum and membrane as well as developing a trajectory energy minimisation function along time-lapse series. Experiments show that by applying this scheme, cell growth and division can be measured automatically. The results show the efficiency of the approach when testing on different datasets while comparing with other existing algorithms. The proposed approach demonstrates great potential for large scale bacterial cell growth analysis.
Rhinovirus infection is associated with the majority of asthma exacerbations. The role of fractalkine in anti-viral (type 1) and pathogenic (type 2) responses to rhinovirus infection in allergic asthma is unknown. To determine whether (1) fractalkine is produced in airway cells and in peripheral blood leucocytes, (2) rhinovirus infection increases production of fractalkine and (3) levels of fractalkine differ in asthmatic compared to non-asthmatic subjects. Fractalkine protein and mRNA levels were measured in bronchoalveolar lavage (BAL) cells and peripheral blood mononuclear cells (PBMCs) from non-asthmatic controls (n = 15) and mild allergic asthmatic (n = 15) subjects. Protein levels of fractalkine were also measured in macrophages polarised ex vivo to give M1 (type 1) and M2 (type 2) macrophages and in BAL fluid obtained from mild (n = 11) and moderate (n = 14) allergic asthmatic and non-asthmatic control (n = 10) subjects pre and post in vivo rhinovirus infection. BAL cells produced significantly greater levels of fractalkine than PBMCs. Rhinovirus infection increased production of fractalkine by BAL cells from non-asthmatic controls (P
Mycobacterium tuberculosis-induced cellular aggregation is essential for granuloma formation and may assist establishment and early spread of M. tuberculosis infection. The M. tuberculosis ESX1 mutant, which has a non-functional type VII secretion system, induced significantly less production of the host macrophage-derived chemokine fractalkine (CX3CL1). Upon infection of human macrophages ESX1-dependent fractalkine production mediated selective recruitment of CD11b+ monocytic cells and increased infection of neighbouring cells consistent with early local spread of infection. Fractalkine levels were raised in vivo at tuberculous disease sites in humans and were significantly associated with increased CD11b+ monocytic cellular recruitment and extent of granulomatous disease. These findings suggest a novel fractalkine-dependent ESX1-mediated mechanism in early tuberculous disease pathogenesis in humans. Modulation of M. tuberculosis-mediated fractalkine induction may represent a potential treatment option in the future, perhaps allowing us to switch off a key mechanism required by the pathogen to spread between cells. © 2014 The Authors.
Current tuberculosis (TB) vaccine strategies are largely aimed at activating conventional T cell responses to mycobacterial protein antigens. However, the lipid-rich cell wall of Mycobacterium tuberculosis (M. tuberculosis) is essential for pathogenicity and provides targets for unconventional T cell recognition. Group 1 CD1-restricted T cells recognize mycobacterial lipids, but their function in human TB is unclear and their ability to establish memory is unknown. Here, we characterized T cells specific for mycolic acid (MA), the predominant mycobacterial cell wall lipid and key virulence factor, in patients with active TB infection. MA-specific T cells were predominant in TB patients at diagnosis, but were absent in uninfected bacillus Calmette-Guérin-vaccinated (BCG-vaccinated) controls. These T cells were CD1b restricted, detectable in blood and disease sites, produced both IFN-γ and IL-2, and exhibited effector and central memory phenotypes. MA-specific responses contracted markedly with declining pathogen burden and, in patients followed longitudinally, exhibited recall expansion upon antigen reencounter in vitro long after successful treatment, indicative of lipid-specific immunological memory. T cell recognition of MA is therefore a significant component of the acute adaptive and memory immune response in TB, suggesting that mycobacterial lipids may be promising targets for improved TB vaccines.
Cyanobacteria (= blue-green algae) are prolific producers of structurally distinct and biologically active metabolites. In the continuation of our search for new sources of anti-infective natural products, we have assessed the in vitro antiprotozoal (Plasmodium falciparum, Trypanosoma brucei rhodesiense, T. cruzi, Leishmania donovani) and antitubercular (Mycobacterium tuberculosis) potential of samples of two terrestrial cyanobacteria, Nostoc commune (collected when desiccated and wet) and Rivularia biasolettiana. The cytotoxic potential of the extracts was also evaluated against primary L6 cells. Except for T. cruzi and M. tuberculosis, the crude extracts were active against all the organisms tested and showed no toxicity. The crude extracts were then partitioned between n-hexane, chloroform and aqueous methanol and retested against the same panel of pathogens. The chloroform sub-extracts of both N. commune samples showed significant activity against T. b. rhodesiense (IC values 2.0 and 3.5 μg/mL) and P. falciparum (IC s 7.4 and 5.8 μg/mL), with low toxicity. This trend was also true for R. biasolettiana extracts, and its chloroform sub-extract showed notable activity against all parasitic protozoa. There were differences in the biological activity profiles of extracts derived from desiccated and hydrated forms of N. commune. To our knowledge, this is the first study assessing the anti-infective activity of desiccated and hydrated forms of N. commune, as well as R. biasolettiana. Furthermore, the present work reports such biological activity in terrestrial cyanobacteria from Ireland for the first time. These results warrant the further study of Irish terrestrial cyanobacteria as a valuable source of new natural product leads for the treatment of parasitic protozoal infections.