
Dr Kevin Maringer
About
Biography
My research group is working to improve our understanding how the molecular interactions of dengue virus and other flaviviruses with their mosquito vectors contribute to the transmission and global emergence of these viruses. I am passionate about using science to improve health and wellbeing in the real world. I sit on the Microbiology Society policy committee, and have organised training workshops in Indonesia. I am a founding member of the University of Surrey Neglected Tropical Diseases Hub (@NTDhub), an interdisciplinary network bringing together biomedical and social scientists with engineers to solve the biggest challenges in neglected tropical diseases globally. I am also a co-organiser of the Recently Independent Virology Researchers network in the UK. I have developed teaching material for underprivileged youths in New York City, and contributed to programme development at the BBC. My research has been funded by the Wellcome Trust, Medical Research Council, and the Global Challenges Research Fund.
Previously, I studied herpesvirus assembly for my PhD thesis in the lab of Professor Gill Elliott at Imperial College London (London, UK), after graduating with degree in Medical Microbiology and Virology from the University of Warwick (Coventry, UK). I was then funded by the Wellcome Trust to study the molecular interactions between dengue virus and its human host and mosquito vector on a postdoctoral fellowship split between the labs of Professor Ana Fernandez-Sesma (Icahn School of Medicine at Mount Sinai, New York, USA) and Dr. Andrew Davidson (University of Bristol, UK).
Areas of specialism
University roles and responsibilities
- Senior Professional Training Year Tutor (International)
Affiliations and memberships
The Maringer Lab in 2019
 Kevin Maringer
Kevin MaringerThe Maringer Lab in 2017

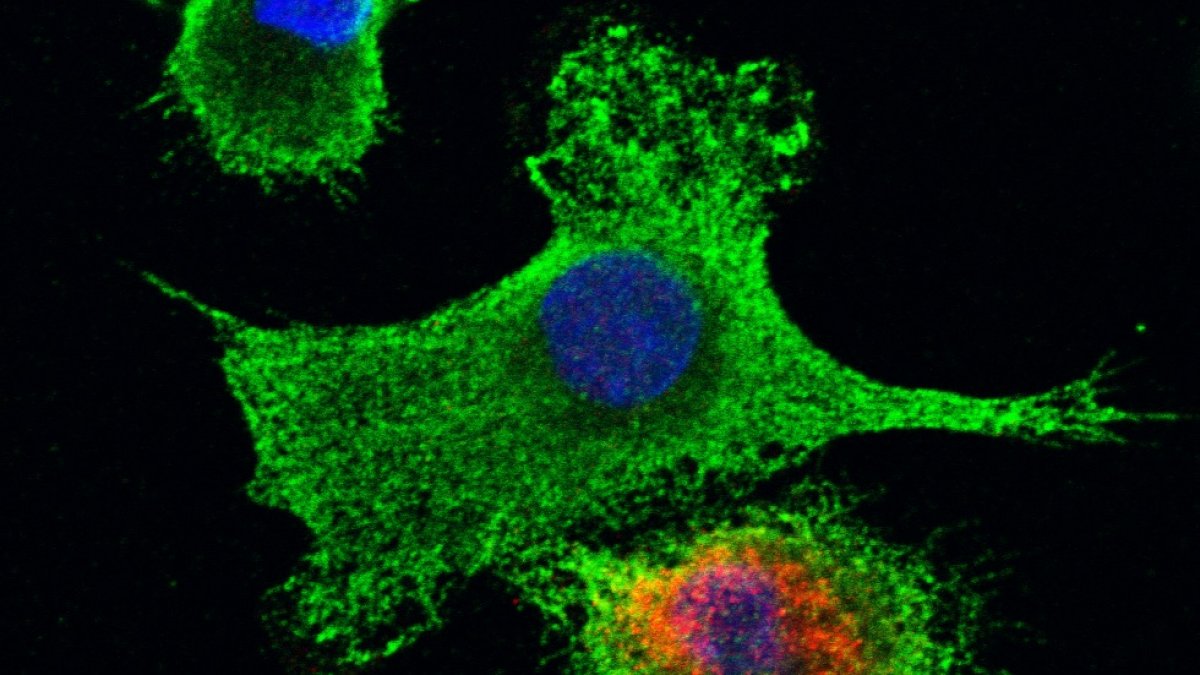
Immune cells infected with dengue virus
Attendees of our 2018 workshop in Makassar, Indonesia
ResearchResearch interests
The Maringer Lab is working to understand the fundamental processes that determine how mosquito-borne viruses are transmitted between humans, alongside studies into the mechanisms that drive the global emergence and spread of new mosquito-borne diseases. We are most interested in the flavivirus dengue virus, which is the most significant arthropod-borne virus (arbovirus) affecting humans, with an estimated 400 million infections across the globe each year. We are also studying the related Zika virus to understand how this virus emerged so rapidly across the Americas. Our research focuses on the mosquito species Aedes aegypti (the 'yellow fever mosquito') and Aedes albopictus (the 'Asian tiger mosquito'), which are distributed globally across the tropics and subtropics and are vectors for dengue virus and Zika virus as well as other important arboviruses that infect humans (for example yellow fever virus and chikungunya virus). Because many mosquito-borne viruses lack effective vaccines or antiviral therapies, targeting their mosquito vectors remains one of our most important strategies for preventing human disease.
Our approach is to use cutting-edge 'omics' technologies to profile global responses of mosquito cells to viral infection. For example, we use 'proteomics' methods to measure every protein in a given sample, which can tell us how mosquito cells respond when infected with dengue or Zika virus (e.g. does the immune system activate?), and whether these viruses alter the cellular environment for their own benefit (e.g. do they steal the cell's nutrients?). We follow up on the big data sets generated using gene editing technologies (CRISPR-Cas9) and molecular tools developed in our lab. By comparing related and unrelated arboviruses in this way, we can pinpoint virus-specific and broadly-applicable molecular mechanisms that drive the transmission, emergence and global spread of flaviviruses like dengue virus and Zika virus. This information will facilitate the development of vector-targeted interventions to reduce the global burden of arboviral disease.
If you are a prospective student or postdoc interested in joining our research group, please get in touch. We welcome ERASMUS students, please get in touch if you are interested in our research.
Research collaborations
We are working with Dr. Paola Campagnolo (School of Bioscience and Medicine) to study how dengue virus causes vascular leakage in haemorrhagic fever patients.
We are collaborating with Dr. Andrew Davidson and Dr. David Matthews (both University of Bristol, UK) to develop new bioinformatic methods to study the proteomes and transcriptomes of mosquito vectors of human disease.
We are collaborating with Dr. Rennos Fragkoudis (Pirbright Institute) and Professor Andres Merits (Institute of Technology, University of Tartu, Estonia) to study molecular drivers of arbovirus emergence in mosquitoes.
We are collaborating with Dr. Jacob Crawford as part of Verily Life Science's "debug" project, which is aimed at improving our understanding of mosquito genetics to prevent human disease.
Research interests
The Maringer Lab is working to understand the fundamental processes that determine how mosquito-borne viruses are transmitted between humans, alongside studies into the mechanisms that drive the global emergence and spread of new mosquito-borne diseases. We are most interested in the flavivirus dengue virus, which is the most significant arthropod-borne virus (arbovirus) affecting humans, with an estimated 400 million infections across the globe each year. We are also studying the related Zika virus to understand how this virus emerged so rapidly across the Americas. Our research focuses on the mosquito species Aedes aegypti (the 'yellow fever mosquito') and Aedes albopictus (the 'Asian tiger mosquito'), which are distributed globally across the tropics and subtropics and are vectors for dengue virus and Zika virus as well as other important arboviruses that infect humans (for example yellow fever virus and chikungunya virus). Because many mosquito-borne viruses lack effective vaccines or antiviral therapies, targeting their mosquito vectors remains one of our most important strategies for preventing human disease.
Our approach is to use cutting-edge 'omics' technologies to profile global responses of mosquito cells to viral infection. For example, we use 'proteomics' methods to measure every protein in a given sample, which can tell us how mosquito cells respond when infected with dengue or Zika virus (e.g. does the immune system activate?), and whether these viruses alter the cellular environment for their own benefit (e.g. do they steal the cell's nutrients?). We follow up on the big data sets generated using gene editing technologies (CRISPR-Cas9) and molecular tools developed in our lab. By comparing related and unrelated arboviruses in this way, we can pinpoint virus-specific and broadly-applicable molecular mechanisms that drive the transmission, emergence and global spread of flaviviruses like dengue virus and Zika virus. This information will facilitate the development of vector-targeted interventions to reduce the global burden of arboviral disease.
If you are a prospective student or postdoc interested in joining our research group, please get in touch. We welcome ERASMUS students, please get in touch if you are interested in our research.
Research collaborations
We are working with Dr. Paola Campagnolo (School of Bioscience and Medicine) to study how dengue virus causes vascular leakage in haemorrhagic fever patients.
We are collaborating with Dr. Andrew Davidson and Dr. David Matthews (both University of Bristol, UK) to develop new bioinformatic methods to study the proteomes and transcriptomes of mosquito vectors of human disease.
We are collaborating with Dr. Rennos Fragkoudis (Pirbright Institute) and Professor Andres Merits (Institute of Technology, University of Tartu, Estonia) to study molecular drivers of arbovirus emergence in mosquitoes.
We are collaborating with Dr. Jacob Crawford as part of Verily Life Science's "debug" project, which is aimed at improving our understanding of mosquito genetics to prevent human disease.
Publications
Dengue virus (DENV) is the most significant arthropod-borne virus (arbovirus) of humans, primarily transmitted by Aedes aegypti mosquitoes. Currently there are no specific therapeutics and the existing vaccine exhibits limited efficacy. Therefore, vector control remains the best approach to manage disease spread. We previously demonstrated that DENV-2 infection does not induce innate immunodeficiency (IMD) signalling in the Ae. aegypti Aag2 cell line, recapitulating in vivo data from other groups. Furthermore, prior infection with DENV-2 reduces IMD signalling activation by classical immune stimuli. This project aimed to identify DENV-2 antagonist(s) responsible for this immune inhibition using an RT-qPCR-based screening platform in which IMD signalling is stimulated in cells expressing DENV-2 proteins individually. Our results identified NS4A as a tentative antagonist, which can now be used to enhance our understanding of Ae. aegypti antiviral immunity by investigating virus-host interactions. The study of vector immunity is hampered by the lack of tools such as antibodies and cell lines. Our group previously created CRISPR-Cas9 knockout Aag2 cell lines, which lack genes essential in the innate immune pathways. These knockout cell lines were created from clonally selected Aag2 cells derived from the heterogeneous parental cell line, and this report also describes the final characterisation of these clones. Results confirm that the cells are embryonic in origin, which confounded our sex analysis. Aag2 clones were confirmed to be persistently infected insect-specific viruses, cell fusing agent virus and Phasi Charoen-like virus. Transfection efficiencies were also determined for the clones of interest. Finally, mutations introduced by CRISPR-Cas9 were characterised in cells derived from one of the clonally selected lines, with one clone identified as the intended mutant, however the IMD pathway-deficient cell clones were determined as wild type. Ultimately insights into vector antiviral immunity may contribute towards development of transmission-incompetent mosquitoes, thereby reducing the global burden of dengue.
Dengue is the most prevalent arthropod-borne viral disease affecting humans, with severe dengue typified by potentially fatal microvascular leakage and hypovolaemic shock. Blood vessels of the microvasculature are composed of a tubular structure of endothelial cells ensheathed by perivascular cells (pericytes). Pericytes support endothelial cell barrier formation and maintenance through paracrine and contact-mediated signalling, and are critical to microvascular integrity. Pericyte dysfunction has been linked to vascular leakage in noncommunicable pathologies such as diabetic retinopathy, but has never been linked to infection-related vascular leakage. Dengue vascular leakage has been shown to result in part from the direct action of the secreted dengue virus (DENV) non-structural protein NS1 on endothelial cells. Using primary human vascular cells, we show here that NS1 also causes pericyte dysfunction, and that NS1-induced endothelial hyperpermeability is more pronounced in the presence of pericytes. Notably, NS1 specifically disrupted the ability of pericytes to support endothelial cell function in a 3D microvascular assay, with no effect on pericyte viability or physiology. These effects are mediated at least in part through contact-independent paracrine signals involved in endothelial barrier maintenance by pericytes. We therefore identify a role for pericytes in amplifying NS1-induced microvascular hyperpermeability in severe dengue, and thus show that pericytes can play a critical role in the aetiology of an infectious vascular leakage syndrome. These findings open new avenues of research for the development of drugs and diagnostic assays for combating infection-induced vascular leakage, such as severe dengue.
Zika virus (ZIKV) is a mosquito borne flavivirus, which was a neglected tropical pathogen until it emerged and spread across the Pacific Area and the Americas, causing large human outbreaks associated with fetal abnormalities and neurological disease in adults. The factors that contributed to the emergence, spread and change in pathogenesis of ZIKV are not understood. We previously reported that ZIKV evades cellular antiviral responses by targeting STAT2 for degradation in human cells. In this study, we demonstrate that Stat2-/- mice are highly susceptible to ZIKV infection, recapitulate virus spread to the central nervous system (CNS), gonads and other visceral organs, and display neurological symptoms. Further, we exploit this model to compare ZIKV pathogenesis caused by a panel of ZIKV strains of a range of spatiotemporal history of isolation and representing African and Asian lineages. We observed that African ZIKV strains induce short episodes of severe neurological symptoms followed by lethality. In comparison, Asian strains manifest prolonged signs of neuronal malfunctions, occasionally causing death of the Stat2-/- mice. African ZIKV strains induced higher levels of inflammatory cytokines and markers associated with cellular infiltration in the infected brain in mice, which may explain exacerbated pathogenesis in comparison to those of the Asian lineage. Interestingly, viral RNA levels in different organs did not correlate with the pathogenicity of the different strains. Taken together, we have established a new murine model that supports ZIKV infection and demonstrate its utility in highlighting intrinsic differences in the inflammatory response induced by different ZIKV strains leading to severity of disease. This study paves the way for the future interrogation of strain-specific changes in the ZIKV genome and their contribution to viral pathogenesis.
Although the herpes simplex virus type 1 (HSV-1) tegument is comprised of a large number of viral and cellular proteins, how and where in the cell these proteins are recruited into the virus structure is poorly understood. We have shown previously that the immediate-early gene product ICP0 is packaged by a mechanism dependent on the major tegument protein VP22, while others have shown a requirement for ICP27. We now extend our studies to show that ICP0 packaging correlates directly with the ability of ICP0 to complex with VP22 in infected cells. ICP27 is not, however, present in this VP22-ICP0 complex but is packaged into the virion in a VP22- and ICP0-independent manner. Biochemical fractionation of virions indicated that ICP0 associates tightly with the virus capsid, but intranuclear capsids contained no detectable ICP0. The RING finger domain of ICP0 and the N terminus of VP22 were both shown to be essential but not sufficient for ICP0 packaging and complex formation. Strikingly, however, the N-terminal region of VP22, while unable to form a complex with ICP0, inhibited its translocation from the nucleus to the cytoplasm. PML degradation by ICP0 was efficient in cells infected with this VP22 mutant virus, confirming that ICP0 retains activity. Hence, we would suggest that VP22 is an important molecular partner of ICP0 that controls at least one of its activities: its assembly into the virion. Moreover, we propose that the pathway by which VP22 recruits ICP0 to the virion may begin in the nucleus prior to ICP0 translocation to its final site of assembly in the cytoplasm. Copyright © 2010, American Society for Microbiology.
The small interfering RNA (siRNA) pathway is a major antiviral response in mosquitoes; however, another RNA interference pathway, the PIWI-interacting RNA (piRNA) pathway, has been suggested to be antiviral in mosquitoes. Piwi4 has been reported to be a key mediator of this response in mosquitoes, but it is not involved in the production of virus-specific piRNAs. Here, we show that Piwi4 associates with members of the antiviral exogenous siRNA pathway (Ago2 and Dcr2), as well as with proteins of the piRNA pathway (Ago3, Piwi5, and Piwi6) in an Aedes aegypti-derived cell line, Aag2. Analysis of small RNAs captured by Piwi4 revealed that it is predominantly associated with virus-specific siRNAs in Semliki Forest virusinfected cells and, to a lesser extent, with viral piRNAs. By using a Dcr2 knockout cell line, we showed directly that Ago2 lost its antiviral activity, as it was no longer bound to siRNAs, but Piwi4 retained its antiviral activity in the absence of the siRNA pathway. These results demonstrate a complex interaction between the siRNA and piRNA pathways in A. aegypti and identify Piwi4 as a noncanonical PIWI protein that interacts with members of the siRNA and piRNA pathways, and its antiviral activities may be independent of either pathway.
Dengue virus (DENV) is a pathogen with a high impact on human health. It replicates in a wide range of cells involved in the immune response. To efficiently infect humans, DENV must evade or inhibit fundamental elements of the innate immune system, namely the type I interferon response. DENV circumvents the host immune response by expressing proteins that antagonize the cellular innate immunity. We have recently documented the inhibition of type I IFN production by the proteolytic activity of DENV NS2B3 protease complex in human monocyte derived dendritic cells (MDDCs). In the present report we identify the human adaptor molecule STING as a target of the NS2B3 protease complex. We characterize the mechanism of inhibition of type I IFN production in primary human MDDCs by this viral factor. Using different human and mouse primary cells lacking STING, we show enhanced DENV replication. Conversely, mutated versions of STING that cannot be cleaved by the DENV NS2B3 protease induced higher levels of type I IFN after infection with DENV. Additionally, we show that DENV NS2B3 is not able to degrade the mouse version of STING, a phenomenon that severely restricts the replication of DENV in mouse cells, suggesting that STING plays a key role in the inhibition of DENV infection and spread in mice.
A large proportion of the genome of most eukaryotic organisms consists of highly repetitive mobile genetic elements. The sum of these elements is called the ‘mobilome’, which in eukaryotes is made up mostly of transposons. Transposable elements contribute to disease, evolution, and normal physiology by mediating genetic rearrangement, and through the ‘domestication’ of transposon proteins for cellular functions. Although ‘omics studies of mobilome genomes and transcriptomes are common, technical challenges have hampered high-throughput global proteomics analyses of transposons. In a recent paper, we overcame these technical hurdles using a technique called ‘proteomics informed by transcriptomics' (PIT), and thus published the first unbiased global mobilome-derived proteome for any organism (using cell lines derived from the mosquito Aedes aegypti). In this commentary, we describe our methods in more detail, and summarise our major findings. We also use new genome sequencing data to show that, in many cases, the specific genomic element expressing a given protein can be identified using PIT. This proteomic technique therefore represents an important technological advance that will open new avenues of research into the role that proteins derived from transposons and other repetitive and sequence diverse genetic elements, such as endogenous retroviruses, play in health and disease.
Assembly of the herpesvirus tegument is poorly understood but is believed to involve interactions between outer tegument proteins and the cytoplasmic domains of envelope glycoproteins. Here, we present the detailed characterization of a multicomponent glycoprotein-tegument complex found in herpes simplex virus 1 (HSV-1)-infected cells. We demonstrate that the tegument protein VP22 bridges a complex between glycoprotein E (gE) and glycoprotein M (gM). Glycoprotein I (gI), the known binding partner of gE, is also recruited into this gE-VP22-gM complex but is not required for its formation. Exclusion of the glycoproteins gB and gD and VP22's major binding partner VP16 demonstrates that recruitment of virion components into this complex is highly selective. The immediate-early protein ICP0, which requires VP22 for packaging into the virion, is also assembled into this gE-VP22-gM-gI complex in a VP22-dependent fashion. Although subcomplexes containing VP22 and ICP0 can be formed when either gE or gM are absent, optimal complex formation requires both glycoproteins. Furthermore, and in line with complex formation, neither of these glycoproteins is individually required for VP22 or ICP0 packaging into the virion, but deletion of gE and gM greatly reduces assembly of both VP22 and ICP0. Double deletion of gE and gM also results in small plaque size, reduced virus yield, and defective secondary envelopment, similar to the phenotype previously shown for pseudorabies virus. Hence, we suggest that optimal gE-VP22-gM-gI-ICP0 complex formation correlates with efficient virus morphogenesis and spread. These data give novel insights into the poorly understood process of tegument acquisition.
The mechanism by which herpesviruses acquire their tegument is not yet clear. One model is that outer tegument proteins are recruited by the cytoplasmic tails of viral glycoproteins. In the case of herpes simplex virus tegument protein VP22, interactions with the glycoproteins gE and gD have been shown. We have previously shown that the C-terminal half of VP22 contains the necessary signal for assembly into the virus. Here, we show that during infection VP22 interacts with gE and gM, as well as its tegument partner VP16. However, by using a range of techniques we were unable to demonstrate VP22 binding to gD. By using pulldown assays, we show that while the cytoplasmic tails of both gE and gM interact with VP22, only gE interacts efficiently with the C-terminal packaging domain of VP22. Furthermore, gE but not gM can recruit VP22 to the Golgi/trans-Golgi network region of the cell in the absence of other virus proteins. To examine the role of the gE-VP22 interaction in infection, we constructed a recombinant virus expressing a mutant VP22 protein with a 14-residue deletion that is unable to bind gE ( gEbind). Coimmunoprecipitation assays confirmed that this variant of VP22 was unable to complex with gE. Moreover, VP22 was no longer recruited to its characteristic cytoplasmic trafficking complexes but exhibited a diffuse localization. Importantly, packaging of this variant into virions was abrogated. The mutant virus exhibited poor growth in epithelial cells, similar to the defect we have observed for a VP22 knockout virus. These results suggest that deletion of just 14 residues from the VP22 protein is sufficient to inhibit binding to gE and hence recruitment to the viral envelope and assembly into the virus, resulting in a growth phenotype equivalent to that produced by deleting the entire reading frame.
Viral protein homeostasis depends entirely on the machinery of the infected cell. Accordingly, viruses can illuminate the interplay between cellular proteostasis components and their distinct substrates. Here, we define how the Hsp70 chaperone network mediates the dengue virus life cycle. Cytosolic Hsp70 isoforms are required at distinct steps of the viral cycle, including entry, RNA replication, and virion biogenesis. Hsp70 function at each step is specified by nine distinct DNAJ cofactors. Of these, DnaJB11 relocalizes to virus-induced replication complexes to promote RNA synthesis, while DnaJB6 associates with capsid protein and facilitates virion biogenesis. Importantly, an allosteric Hsp70 inhibitor, JG40, potently blocks infection of different dengue serotypes in human primary blood cells without eliciting viral resistance or exerting toxicity to the host cells. JG40 also blocks replication of other medically-important flaviviruses including yellow fever, West Nile and Japanese encephalitis viruses. Thus, targeting host Hsp70 subnetworks provides a path for broad-spectrum antivirals.
Zika virus (ZIKV) is a mosquito borne flavivirus, which was a neglected tropical pathogen until it emerged and spread across the Pacific Area and the Americas, causing large human outbreaks associated with fetal abnormalities and neurological disease in adults. The factors that contributed to the emergence, spread and change in pathogenesis of ZIKV are not understood. We previously reported that ZIKV evades cellular antiviral responses by targeting STAT2 for degradation in human cells. In this study, we demonstrate that Stat2-/- mice are highly susceptible to ZIKV infection, recapitulate virus spread to the central nervous system (CNS), gonads and other visceral organs, and display neurological symptoms. Further, we exploit this model to compare ZIKV pathogenesis caused by a panel of ZIKV strains of a range of spatiotemporal history of isolation and representing African and Asian lineages. We observed that African ZIKV strains induce short episodes of severe neurological symptoms followed by lethality. In comparison, Asian strains manifest prolonged signs of neuronal malfunctions, occasionally causing death of the Stat2-/- mice. African ZIKV strains induced higher levels of inflammatory cytokines and markers associated with cellular infiltration in the infected brain in mice, which may explain exacerbated pathogenesis in comparison to those of the Asian lineage. Interestingly, viral RNA levels in different organs did not correlate with the pathogenicity of the different strains. Taken together, we have established a new murine model that supports ZIKV infection and demonstrate its utility in highlighting intrinsic differences in the inflammatory response induced by different ZIKV strains leading to severity of disease. This study paves the way for the future interrogation of strain-specific changes in the ZIKV genome and their contribution to viral pathogenesis.
Dengue virus (DENV) is the most prevalent mosquito-borne virus causing human disease. Of the 4 DENV serotypes, epidemiological data suggest that DENV-2 secondary infections are associated with more severe disease than DENV-4 infections. Mass cytometry by time-of-flight (CyTOF) was used to dissect immune changes induced by DENV-2 and DENV-4 in human DCs, the initial targets of primary infections that likely affect infection outcomes. Strikingly, DENV-4 replication peaked earlier and promoted stronger innate immune responses, with increased expression of DC activation and migration markers and increased cytokine production, compared with DENV-2. In addition, infected DCs produced higher levels of inflammatory cytokines compared with bystander DCs, which mainly produced IFN-induced cytokines. These high-dimensional analyses during DENV-2 and DENV-4 infections revealed distinct viral signatures marked by different replication strategies and antiviral innate immune induction in DCs, which may result in different viral fitness, transmission, and pathogenesis.
STING has emerged in recent years as an important signalling adaptor in the activation of type I interferon responses during infection with DNA viruses and bacteria. An increasing body of evidence suggests that STING also modulates responses to RNA viruses, though the mechanisms remain less clear. In this review, we give a brief overview of the ways in which STING facilitates sensing of RNA viruses. These include modulation of RIG-I-dependent responses through STING's interaction with MAVS, and more speculative mechanisms involving the DNA sensor cGAS and sensing of membrane remodelling events. We then provide an in-depth literature review to summarise the known mechanisms by which RNA viruses of the families Flaviviridae and Coronaviridae evade sensing through STING. Our own work has shown that the NS2B/3 protease complex of the flavivirus dengue virus binds and cleaves STING, and that an inability to degrade murine STING may contribute to host restriction in this virus. We contrast this to the mechanism employed by the distantly related hepacivirus hepatitis C virus, in which STING is bound and inactivated by the NS4B protein. Finally, we discuss STING antagonism in the coronaviruses SARS coronavirus and human coronavirus NL63, which disrupt K63-linked polyubiquitination and dimerisation of STING (both of which are required for STING-mediated activation of IRF-3) via their papain-like proteases. We draw parallels with less-well characterised mechanisms of STING antagonism in related viruses, and place our current knowledge in the context of species tropism restrictions that potentially affect the emergence of new human pathogens.
RNA interference (RNAi) controls arbovirus infections in mosquitoes. Two different RNAi pathways are involved in antiviral responses: the PIWI-interacting RNA (piRNA) and exogenous short interfering RNA (exo-siRNA) pathways, which are characterized by the production of virus-derived small RNAs of 25–29 and 21 nucleotides, respectively. The exo-siRNA pathway is considered to be the key mosquito antiviral response mechanism. In Aedes aegypti-derived cells, Zika virus (ZIKV)-specific siRNAs were produced and loaded into the exo-siRNA pathway effector protein Argonaute 2 (Ago2); although the knockdown of Ago2 did not enhance virus replication. Enhanced ZIKV replication was observed in a Dcr2-knockout cell line suggesting that the exo-siRNA pathway is implicated in the antiviral response. Although ZIKV-specific piRNA-sized small RNAs were detected, these lacked the characteristic piRNA ping-pong signature motif and were bound to Ago3 but not Piwi5 or Piwi6. Silencing of PIWI proteins indicated that the knockdown of Ago3, Piwi5 or Piwi6 did not enhance ZIKV replication and only Piwi4 displayed antiviral activity. We also report that the expression of ZIKV capsid (C) protein amplified the replication of a reporter alphavirus; although, unlike yellow fever virus C protein, it does not inhibit the exo-siRNA pathway. Our findings elucidate ZIKV-mosquito RNAi interactions that are important for understanding its spread.
Background Aedes aegypti is a vector mosquito of major public health importance, transmitting arthropod-borne viruses (arboviruses) such as chikungunya, dengue, yellow fever and Zika viruses. Wild mosquito populations are persistently infected at high prevalence with insect-specific viruses that do not replicate in vertebrate hosts. In experimental settings, acute infections with insect-specific viruses have been shown to modulate arbovirus infection and transmission in Ae. aegypti and other vector mosquitoes. However, the impact of persistent insect-specific virus infections, which arboviruses encounter more commonly in nature, has not been investigated extensively. Cell lines are useful models for studying virus-host interactions, however the available Ae. aegypti cell lines are poorly defined and heterogenous cultures. Methodology/Principle findings We generated single cell-derived clonal cell lines from the commonly used Ae. aegypti cell line Aag2. Two of the fourteen Aag2-derived clonal cell lines generated harboured markedly and consistently reduced levels of the insect-specific bunyavirus Phasi Charoen-like virus (PCLV) known to persistently infect Aag2 cells. In contrast to studies with acute insect-specific virus infections in cell culture and in vivo, we found that pre-existing persistent PCLV infection had no major impact on the replication of the flaviviruses dengue virus and Zika virus, the alphavirus Sindbis virus, or the rhabdovirus vesicular stomatitis virus. We also performed a detailed characterisation of the morphology, transfection efficiency and immune status of our Aag2-derived clonal cell lines, and have made a clone that we term Aag2-AF5 available to the research community as a well-defined cell culture model for arbovirus-vector interaction studies. Conclusions/Significance Our findings highlight the need for further in vivo studies that more closely recapitulate natural arbovirus transmission settings in which arboviruses encounter mosquitoes harbouring persistent rather than acute insect-specific virus infections. Furthermore, we provide the well-characterised Aag2-derived clonal cell line as a valuable resource to the arbovirus research community.
Background: Aedes aegypti is a vector for the (re-)emerging human pathogens dengue, chikungunya, yellow fever and Zika viruses. Almost half of the Ae. aegypti genome is comprised of transposable elements (TEs). Transposons have been linked to diverse cellular processes, including the establishment of viral persistence in insects, an essential step in the transmission of vector-borne viruses. However, up until now it has not been possible to study the overall proteome derived from an organism’s mobile genetic elements, partly due to the highly divergent nature of TEs. Furthermore, as for many non-model organisms, incomplete genome annotation has hampered proteomic studies on Ae. aegypti. Results: We analysed the Ae. aegypti proteome using our new proteomics informed by transcriptomics (PIT) technique, which bypasses the need for genome annotation by identifying proteins through matched transcriptomic (rather than genomic) data. Our data vastly increase the number of experimentally confirmed Ae. aegypti proteins. The PIT analysis also identified hotspots of incomplete genome annotation, and showed that poor sequence and assembly quality do not explain all annotation gaps. Finally, in a proof-of principle study, we developed criteria for the characterisation of proteomically active TEs. Protein expression did not correlate with a TE’s genomic abundance at different levels of classification. Most notably, long terminal repeat (LTR) retrotransposons were markedly enriched compared to other elements. PIT was superior to ‘conventional’ proteomic approaches in both our transposon and genome annotation analyses. Conclusions: We present the first proteomic characterisation of an organism’s repertoire of mobile genetic elements, which will open new avenues of research into the function of transposon proteins in health and disease. Furthermore, our study provides a proof-of concept that PIT can be used to evaluate a genome’s annotation to guide annotation efforts which has the potential to improve the efficiency of annotation projects in non-model organisms. PIT therefore represents a valuable new tool to study the biology of the important vector species Ae. aegypti, including its role in transmitting emerging viruses of global public health concern.